Twisted Bilayer Molybdenum Disulfide Structure Creation.¶
Introduction.¶
This tutorial demonstrates the process of creating a twisted bilayer molybdenum disulfide (MoS2) structure based on the work presented in the following manuscript.
Manuscript
Kaihui Liu, Liming Zhang, Ting Cao, Chenhao Jin, Diana Qiu, Qin Zhou, Alex Zettl, Peidong Yang, Steve G. Louie & Feng Wang, "Evolution of interlayer coupling in twisted molybdenum disulfide bilayers" Nature Communications volume 5, Article number: 4966 (2014) DOI: 10.1038/ncomms5966 12
We use the Materials Designer to create molybdenum disulfide bilayer structure configurations with multiple twist angles.
The Figure 4 shows the twisted bilayer MoS2 configurations.

1. Load and preview MoS2 structure.¶
First, we navigate to Materials Designer and import the MoS2 material from the Standata.
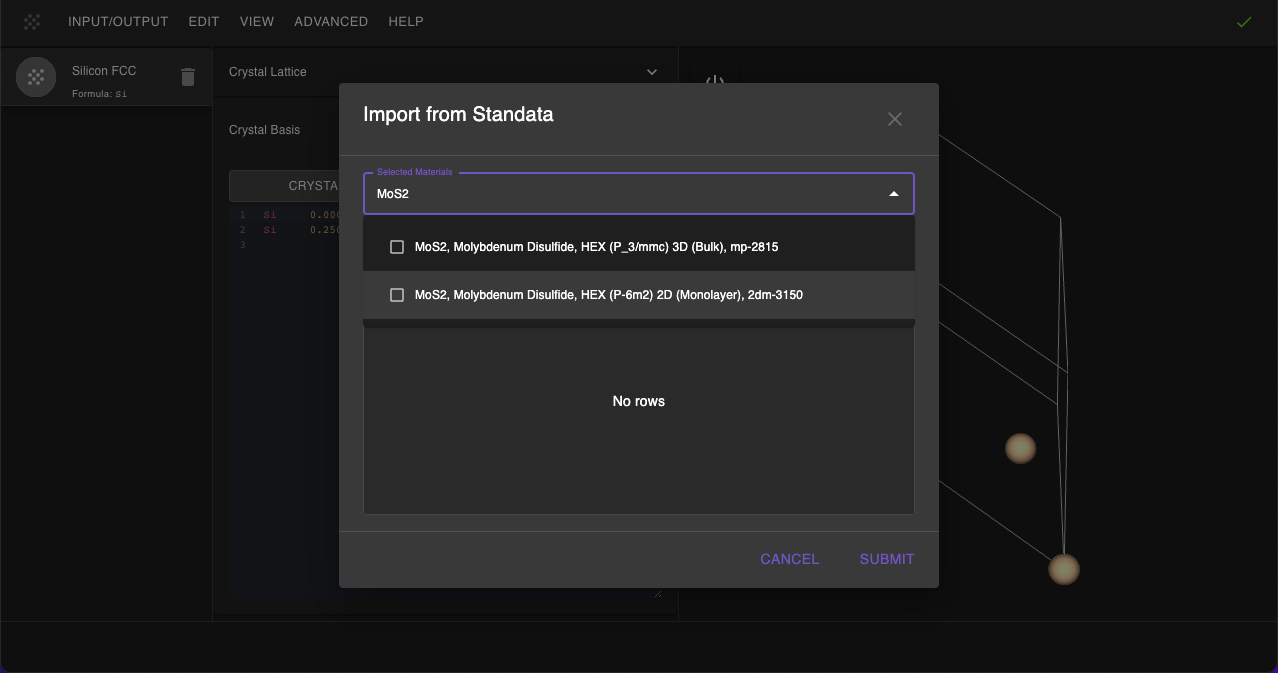
Then we will use the JupyterLite environment to create a twisted bilayer molybdenum disulfide structure.
2. Create MoS2 bilayer with a twist angle of 22 degrees.¶
2.1 Launch JupyterLite Session.¶
Select the "Advanced > JupyterLite Transformation" menu item to launch the JupyterLite environment.
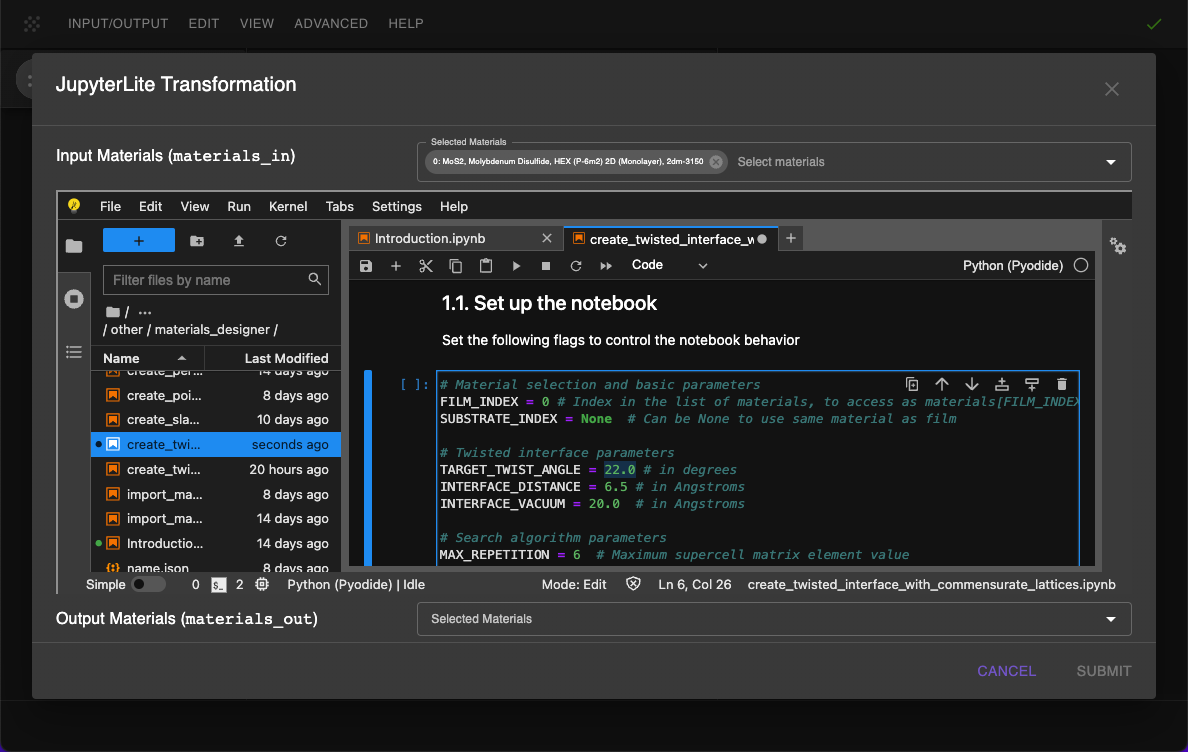
2.2. Open and modify the notebook.¶
Next, edit create_twisted_interface_with_commnesurate_lattices.ipynb notebook to modify the parameters by adding: TARGET_TWIST_ANGLE = 22 and INTERFACE_DISTANCE = 6.5 -- found in the publication description.
Adjust the "1.1. Set up slab parameters" cell in the notebook according to:
# Material selection and basic parameters
FILM_INDEX = 0 # Index in the list of materials, to access as materials[FILM_INDEX]
SUBSTRATE_INDEX = None # Can be None to use same material as film
# Twisted interface parameters
TARGET_TWIST_ANGLE = 22.0 # in degrees
INTERFACE_DISTANCE = 6.5 # in Angstroms
INTERFACE_VACUUM = 20.0 # in Angstroms
# Search algorithm parameters
MAX_REPETITION = 6 # Maximum supercell matrix element value
ANGLE_TOLERANCE = 0.5 # in degrees
RETURN_FIRST_MATCH = True # If True, returns first solution within tolerance
# Visualization parameters
SHOW_INTERMEDIATE_STEPS = True
VISUALIZE_REPETITIONS = [3, 3, 1]
2.3. Run the Notebook.¶
After setting the parameters, run the notebook to create the twisted bilayer molybdenum disulfide structure.
2.4. View Results and pass to Materials Designer.¶
The generation might take some time. After that, the user can pass the material to the Materials Designer for further analysis.
The interface for 22 degrees twist is shown below.
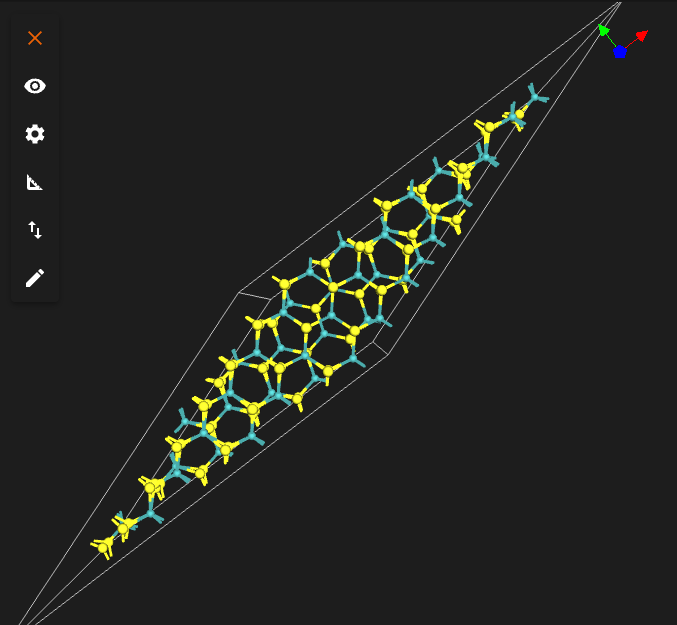
3. Create bilayers with other twist angles.¶
3.1. Repeat the steps above.¶
To create a twisted bilayer MoS2 structure with a different twist angle, repeat the steps above, adjusting the TARGET_TWIST_ANGLE and INTERFACE_DISTANCE parameters accordingly.
Values for angle and associated interlayer separation provided below come from the description of Figure 4 in the publication, below each example has an image of the resulting material.
TARGET_TWIST_ANGLE = 0.0
INTERFACE_DISTANCE = 6.8
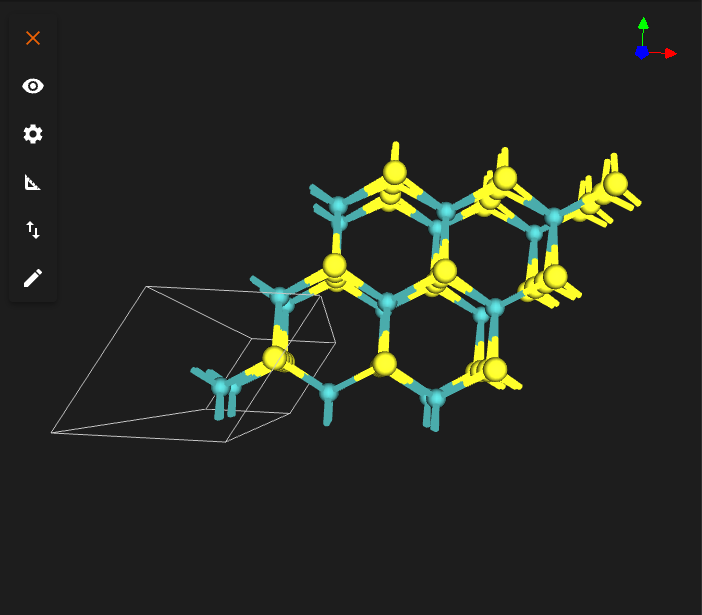
TARGET_TWIST_ANGLE = 13.0
INTERFACE_DISTANCE = 6.5
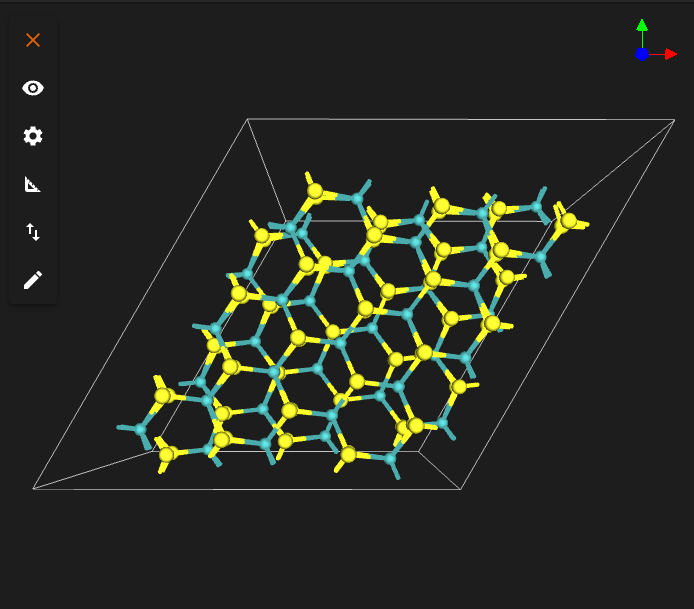
TARGET_TWIST_ANGLE = 38.0
INTERFACE_DISTANCE = 6.5
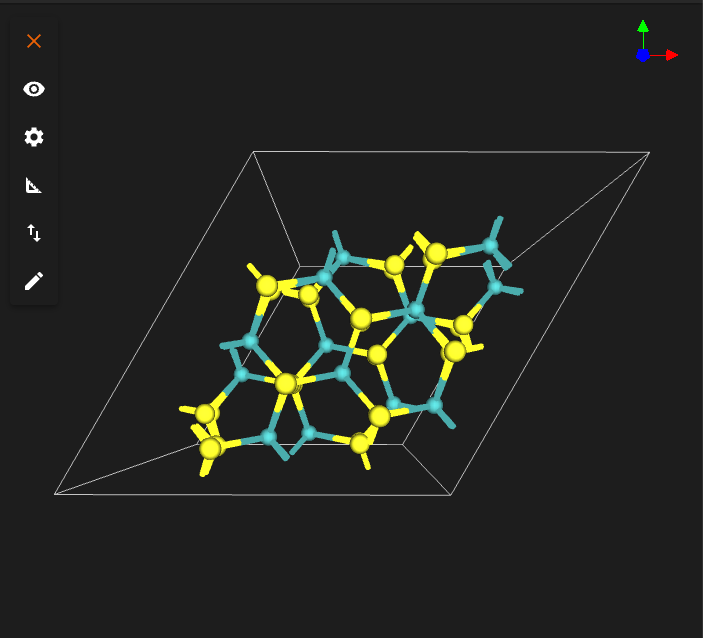
TARGET_TWIST_ANGLE = 47.0
INTERFACE_DISTANCE = 6.5
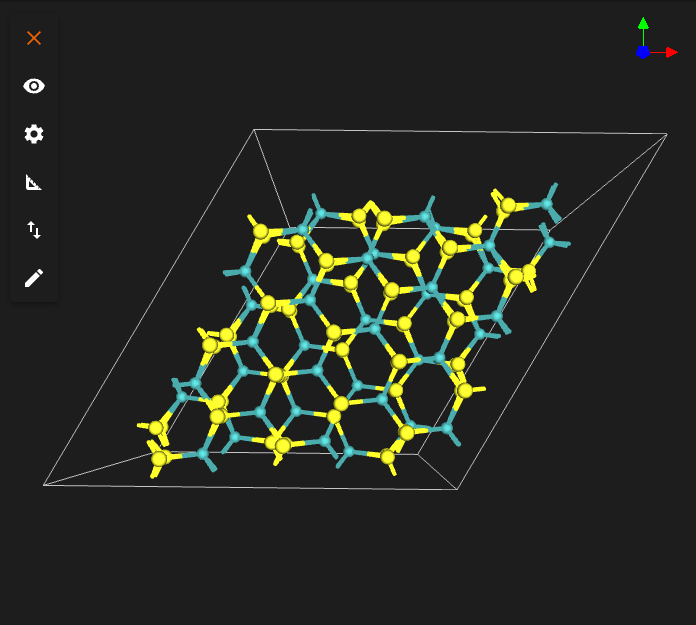
TARGET_TWIST_ANGLE = 60.0
INTERFACE_DISTANCE = 6.2
Interactive JupyterLite Notebook.¶
The interactive JupyterLite notebook for creating twisted bilayer MoS2 structures can be accessed below. To run the notebook, click on the "Run All" button.
References.¶
-
Kaihui Liu, Liming Zhang, Ting Cao, Chenhao Jin, Diana Qiu, Qin Zhou, Alex Zettl, Peidong Yang, Steve G. Louie, and Feng Wang. Evolution of interlayer coupling in twisted molybdenum disulfide bilayers. Nature Communications, 5:4966, 2014. URL: https://doi.org/10.1038/ncomms5966, doi:10.1038/ncomms5966. ↩
-
Y. Cao, V. Fatemi, S. Fang, and et al. Unconventional superconductivity in magic-angle graphene superlattices. Nature, 556:43–50, 2018. URL: https://doi.org/10.1038/nature26160. ↩